What is PharmacopeAI?
PharmacopeAI is a platform that identifies correlations between a patient's unique combination of cancer mutations and responses to anti-cancer drugs.
Who built it?
PharmacopeAI is a joint initiative between the Cancer Science Institute, School of Computing from the National University of Singapore and the National University Cancer Institute Singapore.
Who is it for?
Cancer physicians exploring off-label therapeutic options based on genomics, for patients with cancer refractory to standard treatment.




Cancer
doesn't wait.
There is more info in clinical NGS than what we currently use. Our goal is to address this problem with a practical “what can we do with what we have, right here and right now?” approach.
It is common practice in oncology to order a clinical-grade next-generation sequencing test for patients whose cancers have relapsed after standard treatment, with the hope of identifying a “targetable” genetic alteration. However, only 5% of patients who undergo a clinical NGS test will have matched therapy or matched clinical trials available. This is because current matching is done on a “single genetic mutation- single drug” basis (monogenic prediction).
What can we do for the remaining 95% who are usually fit for treatment, but have no standard options?
What can we do for the remaining 95% who are usually fit for treatment, but have no standard options?Polygenic NGS interpretation for cancer.
Our solution harnesses the principle that the combination of genetic changes in any given cancer sample reflects underlying cellular characteristics- which in turn associate with sensitivity to specific drugs (polygenic prediction). We aim to identify potential therapeutic options for the “untargetable 95%” of cases, using comprehensive (polygenic) interpretation of clinical NGS reports.
Tumour-agnostic repurposing of cancer drugs, using molecular profiles
There is a growing wealth of pre-clinical (laboratory based) data on molecular determinants of responses of cancer cell lines and models to specific drugs. It is humanly impossible to know the millions of possible associations between combinations of mutations and drugs, and to apply them in the context of a specific patient’s set of mutations.
Deep interpretation of clinical next-generation sequencing for cancer
The PharmacopeAI method uses a combination of neural networks and graph databases to scour these databases for drugs that consistently associate with the unique combination of mutations for a given patient.
Enhancing molecular tumour boards
with deep learning
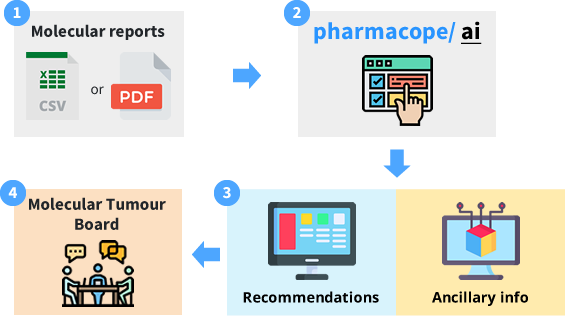
The PharmacopeAI platform is designed to be integrated into current molecular tumour board workflows. The input does not need anything other than a routine clinical NGS report in PDF format (no requirement for raw/ complex data files). The output is currently tailored to focus on inexpensive anti-cancer agents that are in routine clinical use. The medical oncologists in the molecular tumour board can assess the suggestions from the algorithm for suitability for the given patient.
Given that each cancer is a unique conglomerate of mutations, it follows that therapy for each person needs to be unique.